aka ‘CaLibrating and Improving Models of Biodiversity‘.
This project has received funding from the European Union’s
Horizon 2020 research and innovation programme under grant agreement No 840946 as an Individual Marie-Curie Fellowship in 2019 (after four unsuccessful trials – keep faith).
The question
To model community structure or dynamics, we often wish we could use those neat theoretical models that include the fundamental process of population dynamics, such as growth, mortality and species interactions. Think of your classical model by Lotka and Volterra.
Pi is the abundance/biomass of species i, ri its relative growth rate in a given environment and aij its interaction coefficient with species j.
It is clean, it is simple, it reminds us of Ecology 1.01.
But now, think of your favorite ecosystem, how many species co-exist in it? And how many species parameters ri and aij would you have to measure to calibrate this model and make it relevant? Probably way too many.
The solution (?)
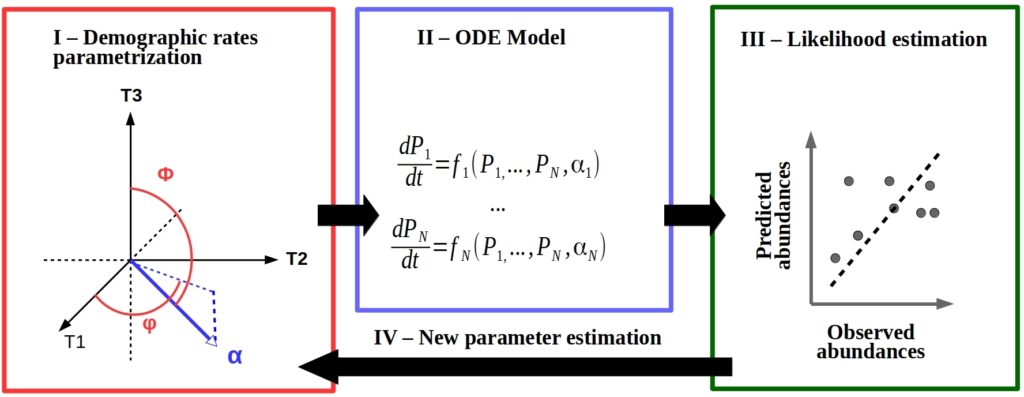
But what if we could use data that were a reasonable proxy for those unknown species parameters that we definitely don’t want to measure? Functional traits are a good candidate. They are linked to species ecological niches and they are fairly easy to gather and can often been found in open databases.
Then the question becomes ‘how to correctly link the species parameters from your favorite community model to the available data on their real-life functional traits?’. A more methodological question that can hopefully be solved with the right statistical framework.
Trait data collection
To complete existing trait databases, we – with Martin Philippe – conducted a field sampling campaign targeting tussock grassland species leaf and root functional traits. The trait sampling took place at the Cass Mountain Research Area.








